source("../../solutions/simple.R")
doomsday_model <- function() {
x = simulate(Unif(0, 5))
observe(0.06, Unif(0, x))
return(x)
}
posterior(doomsday_model, 100000)[1] 1.103447We will write many Stan models in the coming weeks, so we will cover here the key concepts needed to write in Stan. Similarities with simPPLe will help in this process.
Let us revisit the Doomsday model, which we wrote in simPPLe as follows:

source("../../solutions/simple.R")
doomsday_model <- function() {
x = simulate(Unif(0, 5))
observe(0.06, Unif(0, x))
return(x)
}
posterior(doomsday_model, 100000)[1] 1.103447Recall the analytic answer that we computed in clicker questions was: \(\approx 1.117\).
Today we will re-write this model in Stan instead of simPPLe. The main difference is that:
simulate and observe will be denoted by ~ in Stan,data or parameters).First, load rstan:
suppressPackageStartupMessages(require(rstan))Then, as you did to test your install put the following code into a .stan file:
1data {
real y;
}
2parameters {
real<lower=0, upper=5> x;
}
model {
3 x ~ uniform(0, 5);
4 y ~ uniform(0, x);
}data block will be treated as observed.
parameters block will be treated as latent, i.e., unobserved.x is declared latent (i.e., inside a parameters block), Stan will know to treat this as the counterpart of simPPLe’s x = simulate(Unif(0, 5)).
y is declared observed (i.e., inside a data block), Stan will know to treat this as the counterpart of simPPLe’s observe(0.06, Unif(0, x)).
Question: How does Stan get the actual observed value \(y = 0.06\)?
Answer: When Stan gets called in R:1
SAMPLING FOR MODEL 'anon_model' NOW (CHAIN 1).
Chain 1:
Chain 1: Gradient evaluation took 4e-06 seconds
Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 0.04 seconds.
Chain 1: Adjust your expectations accordingly!
Chain 1:
Chain 1:
Chain 1: Iteration: 1 / 100000 [ 0%] (Warmup)
Chain 1: Iteration: 10000 / 100000 [ 10%] (Warmup)
Chain 1: Iteration: 20000 / 100000 [ 20%] (Warmup)
Chain 1: Iteration: 30000 / 100000 [ 30%] (Warmup)
Chain 1: Iteration: 40000 / 100000 [ 40%] (Warmup)
Chain 1: Iteration: 50000 / 100000 [ 50%] (Warmup)
Chain 1: Iteration: 50001 / 100000 [ 50%] (Sampling)
Chain 1: Iteration: 60000 / 100000 [ 60%] (Sampling)
Chain 1: Iteration: 70000 / 100000 [ 70%] (Sampling)
Chain 1: Iteration: 80000 / 100000 [ 80%] (Sampling)
Chain 1: Iteration: 90000 / 100000 [ 90%] (Sampling)
Chain 1: Iteration: 100000 / 100000 [100%] (Sampling)
Chain 1:
Chain 1: Elapsed Time: 0.253 seconds (Warm-up)
Chain 1: 0.157 seconds (Sampling)
Chain 1: 0.41 seconds (Total)
Chain 1: We can now compare the approximation provided by Stan:
fitInference for Stan model: anon_model.
1 chains, each with iter=1e+05; warmup=50000; thin=1;
post-warmup draws per chain=50000, total post-warmup draws=50000.
mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat
x 1.12 0.01 1.25 0.07 0.19 0.55 1.68 4.45 10585 1
lp__ -0.37 0.01 0.63 -2.21 -0.41 -0.12 -0.04 -0.01 9987 1
Samples were drawn using NUTS(diag_e) at Tue Mar 5 11:45:15 2024.
For each parameter, n_eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor on split chains (at
convergence, Rhat=1).;uniform.To access the samples from a “fit” object when calling Stan, use:
samples = extract(fit)
head(samples$x)[1] 0.7224132 3.7228678 2.2768091 1.0077302 4.9788992 0.6378141As a sanity check, let’s make sure all the samples are greater than the observed lower bound of \(0.06\):
min(samples$x)[1] 0.06000474Question: denote the samples output from Stan by \(X^{(1)}, X^{(2)}, \dots, X^{(M)}\). What formula should you use to approximate \(\mathbb{E}[g(X) | Y = y]\) based on the Stan output?
As in simple Monte Carlo, use \(\frac{1}{M} \sum_{m=1}^M g(X^{(m)})\).
Since the samples are equally weighted, this makes it simpler to create plots, e.g., for a histogram:
hist(samples$x)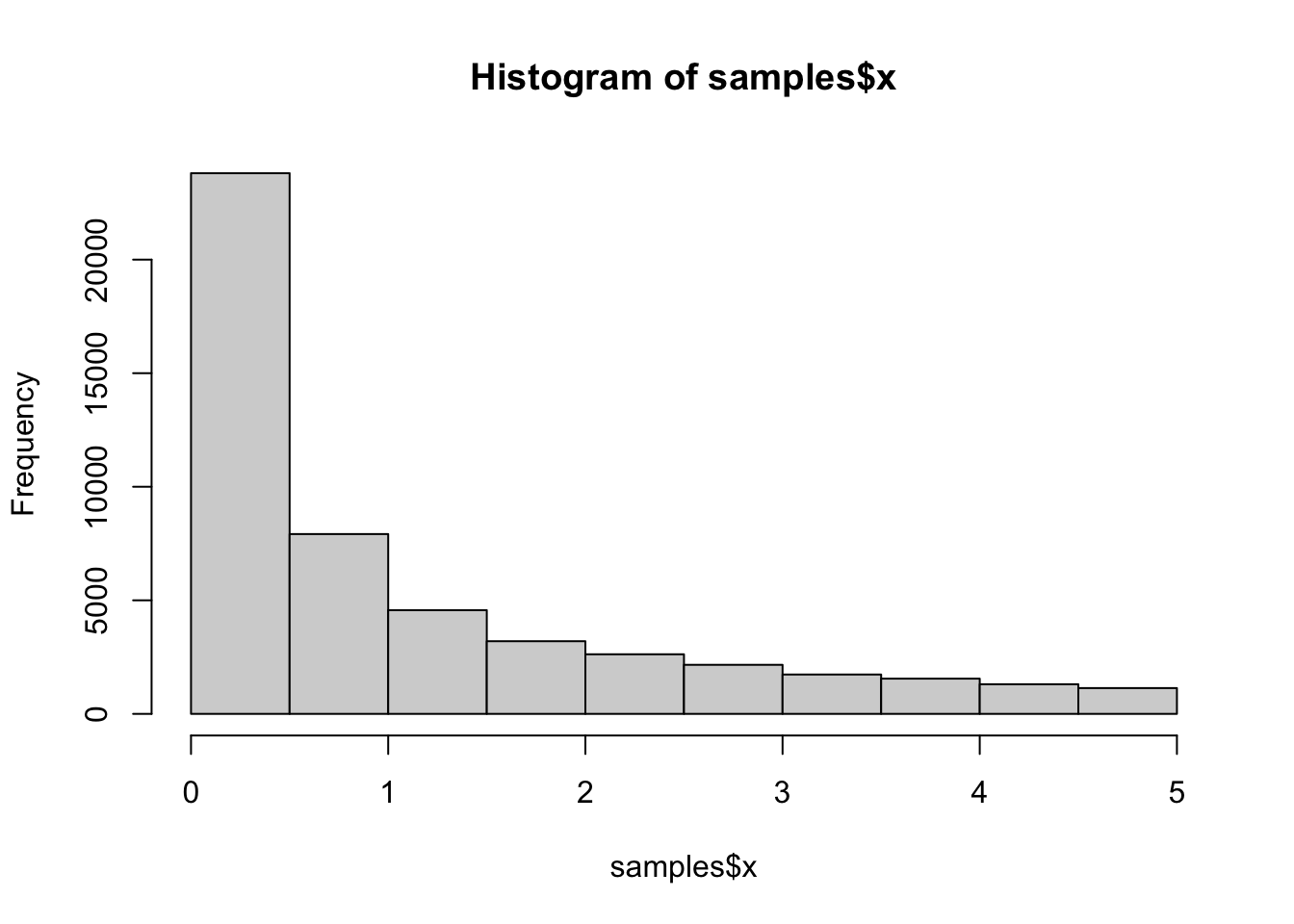
You can also use the bayesplot package which will be handy to create more complex plots from MCMC runs:
suppressPackageStartupMessages(require(bayesplot))
mcmc_hist(fit, pars = c("x"))`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.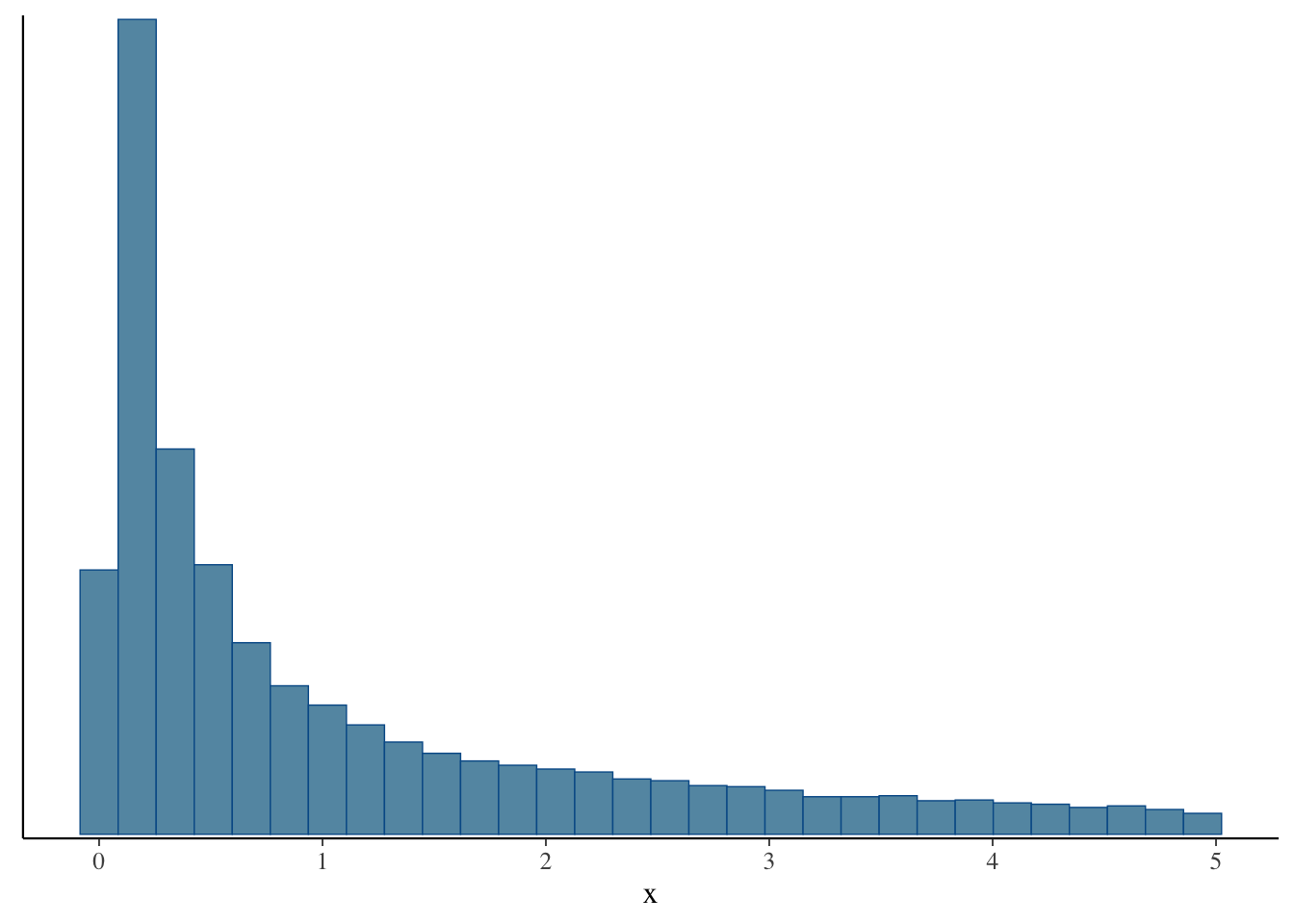
when calling Stan from an R script, use stan(file = ...), but when calling Stan from R Markdown (as we do here), use sampling(name, ...), where the first argument, name matches the code block argument output.var = "name" (see the source of this page for an example).↩︎