suppressPackageStartupMessages(require(rstan))
suppressPackageStartupMessages(require(magrittr))
suppressPackageStartupMessages(require(tidybayes))
suppressPackageStartupMessages(require(ggplot2))Grouped data
Outline
Topics
- Using the package
tidybayesto feed complex data into Stan.
Rationale
Data organized in various groups is a frequent feature of Bayesian models, in particular in hierarchical models.
Getting grouped data into stan is tedious and error prone. The package tidybayes automates much of this.
Pre-reading
If you have never heard about “tidy data”, while it is not strictly essential for this course, it is a good investment to skim this tutorial on tidy data.
Example
First, install the packages magrittr and tidybayes (and ggplot2 if you have not done so already), then import them:
Data prep
We load the data used in exercise 6:
data = read.csv(url("https://raw.githubusercontent.com/UBC-Stat-ML/web447/main/exercises/ex06_assets/vaccines_full.csv"))
data$is_vaccinated = ifelse(data$arms == "vaccinated", 1, 0)
rmarkdown::paged_table(data)The magic conversion of “tidy data” (data in a format like the above) into a format that can be consumed by Stan is done using compose_data:
stan_converted = compose_data(data)
stan_converted$trials
[1] 1 1 3 3 2 2
$n_trials
[1] 3
$arms
[1] 2 1 2 1 2 1
$n_arms
[1] 2
$groupSizes
[1] 5807 5829 18198 18325 14134 14073
$numbersOfCases
[1] 30 101 8 162 11 185
$is_vaccinated
[1] 1 0 1 0 1 0
$n
[1] 6Stan model
Here we consider a simple, non-hierarchical model but fitting all the data at once. In the exercise, you will modify this model to follow the hierarchical structure of exercise 6.
data {
int n;
int n_trials;
array[n] int<lower=1,upper=n_trials> trials;
array[n] int arms;
int n_arms;
array[n] int groupSizes;
array[n] int numbersOfCases;
array[n] int is_vaccinated;
}
parameters {
vector<lower=0,upper=1>[n_trials] efficiencies;
vector<lower=0,upper=1>[n_trials] prevalences;
}
model {
for (trial in 1:n_trials) {
efficiencies[trial] ~ beta(1, 1);
prevalences[trial] ~ beta(1, 1);
}
for (i in 1:n) {
numbersOfCases[i] ~ binomial(groupSizes[i], prevalences[trials[i]] * (is_vaccinated[i] == 1 ? 1.0 - efficiencies[trials[i]] : 1.0));
}
}Fitting
The fit object returned by sampling does not know about the string labels attached to each trial integer index. We use fit %<>% recover_types(data) to add back that information:
fit = sampling(
vaccines,
seed = 1,
data = stan_converted,
refresh = 0,
iter = 10000
)
fit %<>% recover_types(data)Getting draws
The output of Stan is not tidy either.
fitInference for Stan model: anon_model.
4 chains, each with iter=10000; warmup=5000; thin=1;
post-warmup draws per chain=5000, total post-warmup draws=20000.
mean se_mean sd 2.5% 25% 50% 75%
efficiencies[1] 0.69 0.00 0.07 0.55 0.65 0.69 0.73
efficiencies[2] 0.94 0.00 0.02 0.89 0.92 0.94 0.95
efficiencies[3] 0.94 0.00 0.02 0.90 0.93 0.95 0.96
prevalences[1] 0.02 0.00 0.00 0.01 0.02 0.02 0.02
prevalences[2] 0.01 0.00 0.00 0.01 0.01 0.01 0.01
prevalences[3] 0.01 0.00 0.00 0.01 0.01 0.01 0.01
lp__ -2793.24 0.02 1.77 -2797.52 -2794.21 -2792.89 -2791.93
97.5% n_eff Rhat
efficiencies[1] 0.80 22048 1
efficiencies[2] 0.97 23668 1
efficiencies[3] 0.97 24974 1
prevalences[1] 0.02 22889 1
prevalences[2] 0.02 25768 1
prevalences[3] 0.01 25400 1
lp__ -2790.81 8951 1
Samples were drawn using NUTS(diag_e) at Thu Mar 14 17:33:47 2024.
For each parameter, n_eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor on split chains (at
convergence, Rhat=1).Use spread_draws to put the draws into a tidy format:
fit %>% spread_draws(efficiencies[trials], prevalences[trials]) %>% head(10)# A tibble: 10 × 6
# Groups: trials [1]
trials efficiencies .chain .iteration .draw prevalences
<chr> <dbl> <int> <int> <int> <dbl>
1 AZ-Oxford (combined) 0.479 1 1 1 0.0120
2 AZ-Oxford (combined) 0.862 1 2 2 0.0218
3 AZ-Oxford (combined) 0.712 1 3 3 0.0194
4 AZ-Oxford (combined) 0.695 1 4 4 0.0163
5 AZ-Oxford (combined) 0.660 1 5 5 0.0164
6 AZ-Oxford (combined) 0.642 1 6 6 0.0188
7 AZ-Oxford (combined) 0.613 1 7 7 0.0176
8 AZ-Oxford (combined) 0.720 1 8 8 0.0150
9 AZ-Oxford (combined) 0.645 1 9 9 0.0150
10 AZ-Oxford (combined) 0.687 1 10 10 0.0164Plotting
Now that we have draws in tidy format, instead of using specialized MCMC plotting libraries we can just use ggplot:
fit %>%
spread_draws(efficiencies[trials]) %>%
ggplot(aes(x = efficiencies, y = trials)) +
stat_halfeye() +
theme_minimal()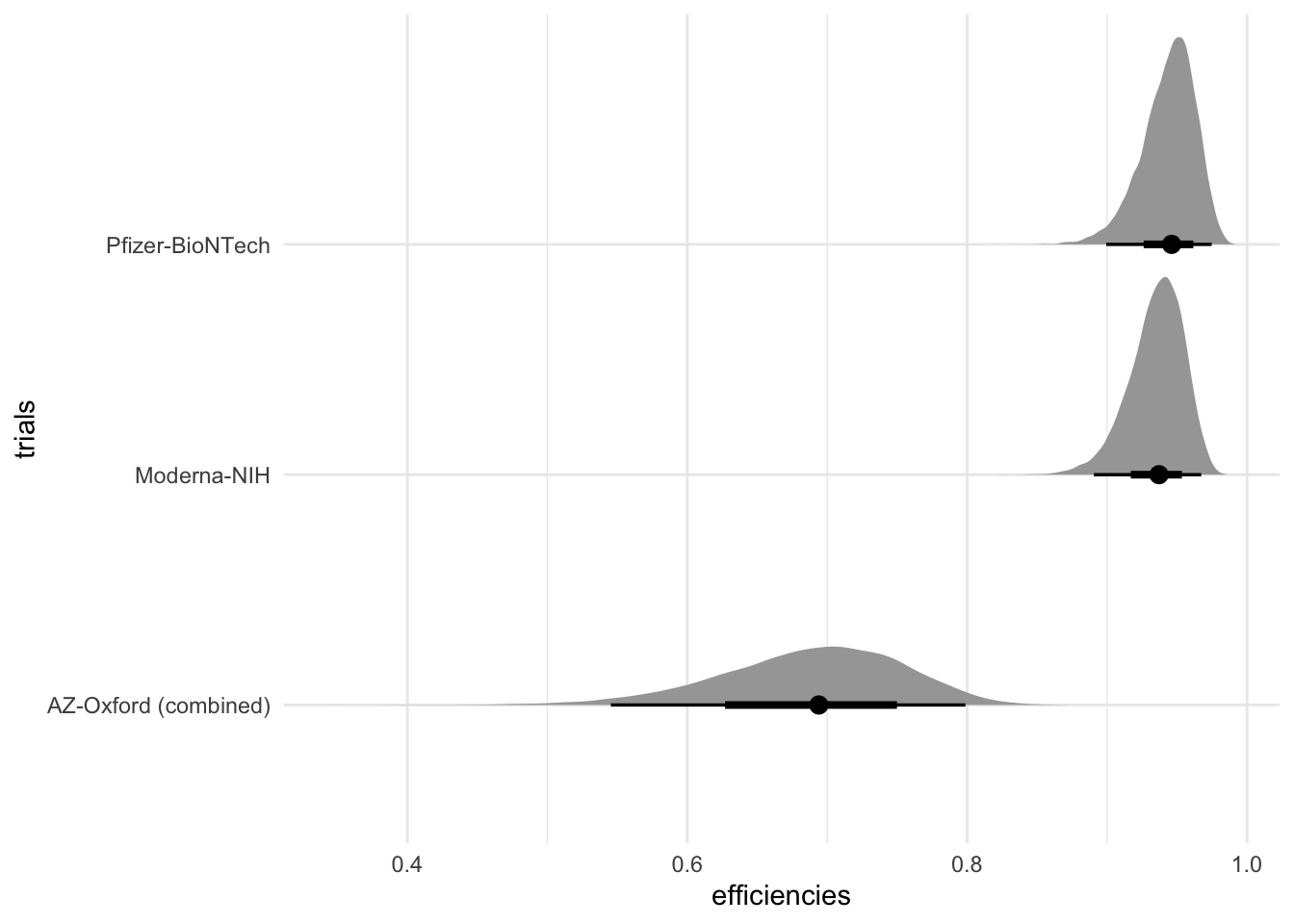
This makes it easier to customize plots.
Summaries
tidybayes also offers convenient ways to compute summaries such as High Density Intervals (HDI)
fit %>%
spread_draws(efficiencies[trials]) %>%
median_hdi(efficiencies)# A tibble: 3 × 7
trials efficiencies .lower .upper .width .point .interval
<chr> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
1 AZ-Oxford (combined) 0.694 0.558 0.806 0.95 median hdi
2 Moderna-NIH 0.937 0.896 0.970 0.95 median hdi
3 Pfizer-BioNTech 0.946 0.905 0.977 0.95 median hdi More information
See the tidybayes documentation.