suppressPackageStartupMessages(require(rstan))
suppressPackageStartupMessages(require(ggplot2))
suppressPackageStartupMessages(require(dplyr))
set.seed(1)
df = read.csv(url("https://raw.githubusercontent.com/UBC-Stat-ML/web447/main/data/launches.csv")) %>% filter(LV.Type == "Ariane 1")
success_indicators = df$Suc_bin
rmarkdown::paged_table(df)Stan: hands on
Outline
We revisit the time-varying Ariane 1 rocket failure probability model to practice the Stan syntax introduced this week.

plot(success_indicators, xlab = "Launch index i")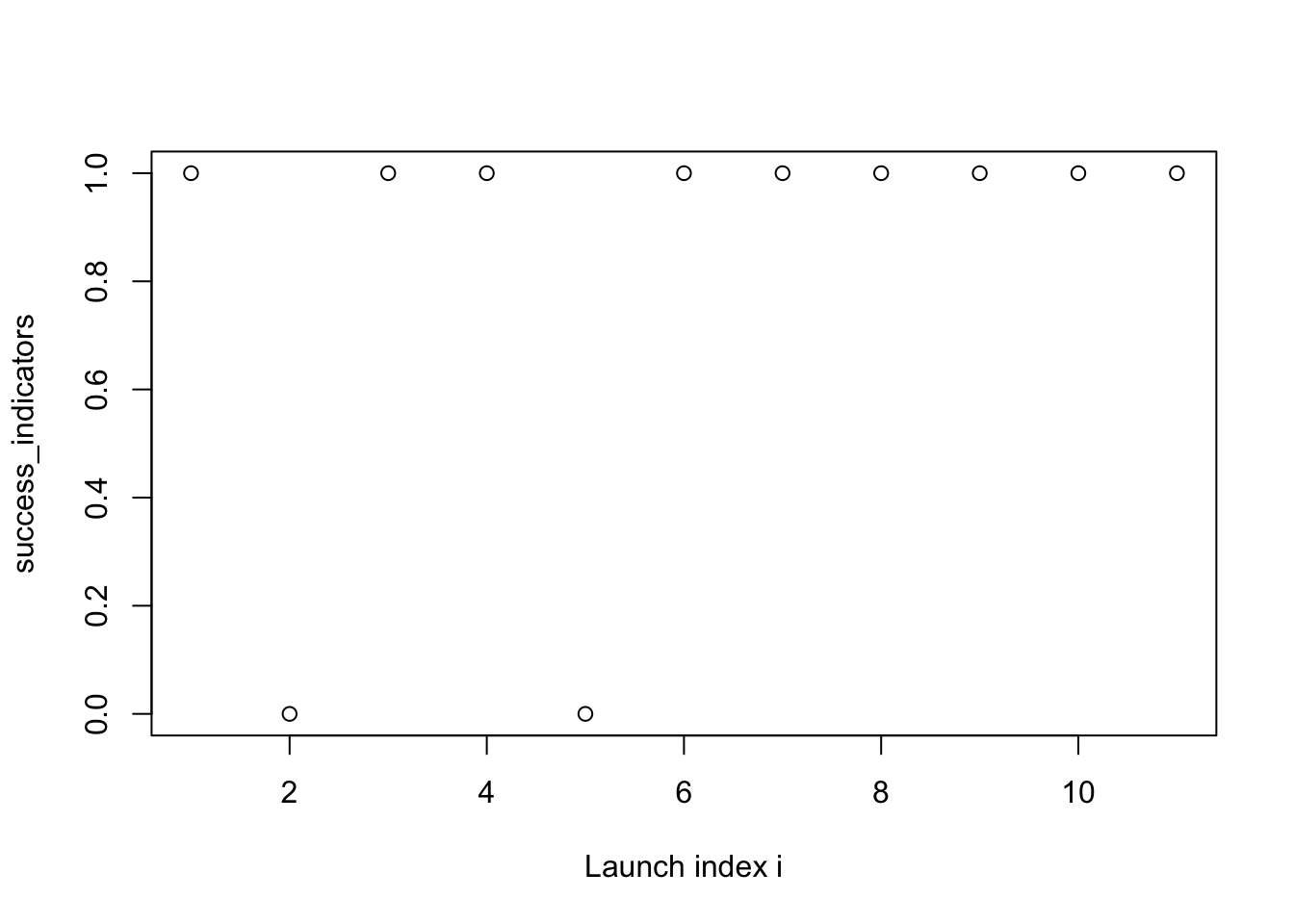
Model
Recall the model we discussed previously (with the difference that we use a SD of 10 now)
\[\begin{align*} \text{slope} &\sim \mathcal{N}(0, 10) \\ \text{intercept} &\sim \mathcal{N}(0, 10) \\ \theta(i) &= \text{logistic}(\text{slope} \cdot i + \text{intercept}) \\ y_i &\sim {\mathrm{Bern}}(\theta(i)) \end{align*}\]
…which you also implemented in simPPLe as part of exercise 4:
logistic_regression = function() {
intercept = simulate(Norm(0, 10))
slope = simulate(Norm(0, 10))
for (i in seq_along(success_indicators)){
success_probability = plogis(intercept + i*slope)
observe(success_indicators[i], Bern(success_probability))
}
return(c(intercept, slope))
}Translation into Stan
Question 1: use the template below to translate the above model into Stan. Set the seed to 1, run 10000 MCMC iterations, and report the posterior mean of the slope parameter.
data {
int N;
array[N] int y;
}
parameters {
real slope;
real intercept;
}
model {
// complete this
}Using the posterior
Question 2: compute a 95% credible interval on the slope parameter.
Use the output of print on the fit object.
Question 3: translate “your updated belief that the Ariane 1 rockets were improving” into a mathematical expression.
Question 4: estimate the numerical value of the expression in the last question.