suppressPackageStartupMessages(require(rstan))Practice questions (Quiz 2)
Page under construction: information on this page may change.
Outline
Topics
- Representative example of questions to prep for Quiz 2.
Important note
Make sure to practice the similar page for Quiz 1 since quiz 2 will cover that material as well.
Libraries needed to run the example below
Model construction
Consider the following setup
- You have a cohort of 15 unemployed persons who are all starting a job search process at the same time.
- For each participant, you have collected 2 covariates: their age and their number of years of education.
- You contact the participants each day for 10 days and record the day they secured a new job.
- At the end of your study, 3 of the participants are still looking for a job.
Using the ~ notation
Define a Bayesian model to handle this dataset. Introduce all random variables, and specify for each its data type, if it observed or not.
Rao-blackwellization
Write the joint density of the model in the last part, before and after Rao-Blackwellization. You can introduce symbols for densities and CDFs, for example leave the density of the exponential as \(p_\text{Exp}(x; \lambda),\) and similarly use \(F_\text{Name}(\cdot; \cdot)\) to denote CDFs.
Model debugging
Let:
- \(C(y)\) denote a 99% credible interval computed from data \(y\).
- Let \(y\) and \(\check y\) denote a real and synthetic (simulated) data respectively.
Suppose you observed the following:
- \(y_n \notin C(y_{\backslash n})\)
- \(\check y_n \notin C(\check y_{\backslash n})\)
- On both synthetic and real data, trace plots and ESS both look good.
- You replicated these experiments several times and always get the same results.
What would you do next? Justify your answer.
Normalization constant
Consider the Bayesian model:
\[\begin{align*} X &\sim {\mathrm{Exp}}(1/100) \\ Y &\sim {\mathrm{Poisson}}(X). \end{align*}\]
When using MCMC, will the output change if, for \(x > 0\)…
- You used \(\check f(x) = \exp(-(1/100) x)\) instead of \(f(x) = (1/100) \exp(-(1/100) x)\) for the prior?
- You used \(\hat f(y|x) = x^y / y!\) instead of \(f(y|x) = \exp(-x) x^y / y!\) for the likelihood?
Intervals
Suppose you see the following output from a Stan MCMC fit object:
Inference for Stan model: anon_model.
1 chains, each with iter=2000; warmup=1000; thin=1;
post-warmup draws per chain=1000, total post-warmup draws=1000.
mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat
slope 0.42 0.00 0.04 0.34 0.40 0.42 0.45 0.51 719 1
sigma 0.24 0.00 0.04 0.18 0.21 0.24 0.27 0.33 639 1
prediction 0.64 0.01 0.27 0.14 0.47 0.64 0.81 1.20 1023 1
lp__ 21.32 0.06 1.04 18.46 20.94 21.62 22.09 22.36 331 1
Samples were drawn using NUTS(diag_e) at Thu Mar 14 14:41:50 2024.
For each parameter, n_eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor on split chains (at
convergence, Rhat=1).- Report a 80% confidence interval to capture the Monte Carlo error for the posterior mean of the
predictionparameter.
Some of the following will be helpful in answering that question:
qnorm(0.8)[1] 0.8416212qnorm(0.9)[1] 1.281552qnorm(0.95)[1] 1.644854- Report a 50% credible interval for the
predictionparameter. You can ignore Monte Carlo error in this sub-question.
Bias and consistency
Let \(\hat G_M\) denote a Monte Carlo estimator based on \(M\) iterations, providing an approximation for an intractable expectation, \(g^* = \mathbb{E}_\pi[g(X)]\).
- Define the notion of bias.
- Define the notion of consistency.
- In the context of Monte Carlo methods, what is more important, a bias of zero (unbiasness) or consistency? Why?
Stan-based prediction
Consider the following code to perform Bayesian linear regression on galaxy distances and velocities:
data {
int<lower=0> N; // number of observations
vector[N] xs; // independent variable
vector[N] ys; // dependent variable
}
parameters {
real slope;
real<lower=0> sigma;
}
model {
// prior
slope ~ student_t(3, 0, 100);
sigma ~ exponential(0.001);
// likelihood
ys ~ normal(slope*xs, sigma);
}How would you modify this code to predict the velocity of a galaxy at distance 1.5? Hint: use the function normal_rng(mean, sd) to generate a normal random variable with the provided mean and standard deviation parameters.
Metropolis-Hastings
Fill the two gaps in the pseudo code below:
- Initialize \(X^{(0)}\) arbitrarily
- For \(m = 1, 2, \dots, M\) do:
- Denote the proposal at iteration \(m \in \{1, 2, \dots, M\}\) by: \[\tilde X^{(m)}\sim q(\cdot | X^{(m-1)}).\]
- Compute the MH ratio: \[R^{(m)}= \frac{\gamma(\tilde X^{(m)})}{\gamma(X^{(m-1)})}.\]
- Sample an acceptance Bernoulli: \[A^{(m)}\sim \text{???}.\]
- If \(A^{(m)}= 1\), we accept the proposed sample: \[X^{(m)}= \text{???}\]
- Else, \(A^{(m)}= 0\), and we reject the proposed sample and stay at previous position: \[X^{(m)}= X^{(m-1)}.\]
Stan
- Explain the difference between the
parametersblock and thetransformed parametersblock in Stan. - Why do you think Stan uses 4 independent chains by default?
Reasoning about MH
Consider the MH algorithm where we use as proposal a normal centered at the current point with standard deviation \(\sigma_p\).
You observe the following trace plot:
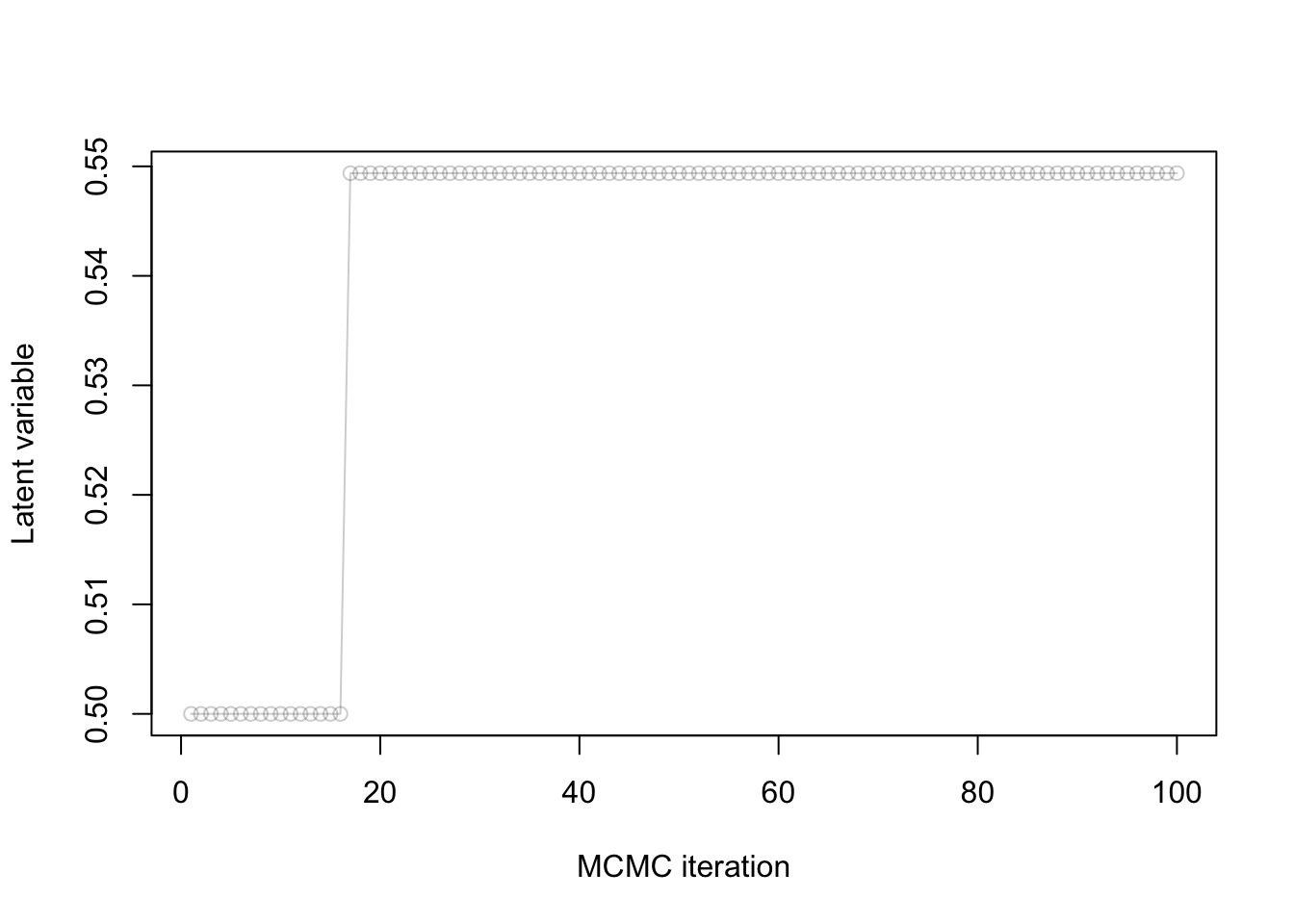
- Is this chain mixing well?
- Why or why not?
- If it is not, what course of action do you recommend?
Irreducibility
Consider the following MH setup:
- \(\gamma(x) = \mathbb{1}[x \in \{1, 2, \dots, 10\}]\)
- \(q(x' | x) = \mathbb{1}[x' \in \{x-1, x+1\}]/2\).
- Define irreducibility.
- Prove that the MH algorithm is irreducible in this setup.
MCMC diagnostics
Explain how to detect slow mixing from a rank plot.
MCSE
How would you proceed if you want to decrease Monte Carlo Standard Error (MCSE)?
Debugging
You wish to write Stan model for logistic regression with normal priors on the parameters with prior variance 100.
Consider the following draft of a Stan model:
data {
int N
array[N] int y
}
model {
slope ~ normal(0, 100);
intercept ~ normal(0, 100);
for (i in 1:N) {
y[i] ~ bernoulli(inv_logit(intercept + slope * i));
}
}Identify as many bugs as you can, and correct each.
Effective sample size from asymptotic variance
Recall that the CLT for i.i.d. and Markov chains give us the following approximations:
- \(\sqrt{M} (\bar X_\text{Markov} - \mu) \approx \sigma_a G,\)
- \(\sqrt{n_e} (\bar X_\text{iid} - \mu) \approx \sigma G,\)
where \(M\) is the number of iterations, \(n_e\) is the effective sample size, \(\bar X_\text{Markov}\) and \(\bar X_\text{iid}\) are the MC estimators based on MCMC and i.i.d. sampling respectively, \(G\) is standard normal, \(\mu\), \(\sigma\) are the posterior mean and standard deviation, and \(\sigma_a\) is the asymptotic variance.
Use these two approximations to write a formula for the effective sample size based on \(\sigma\), \(\sigma_a\) and \(M\).
Independence: prior vs posterior
Suppose we place a prior where \(X_1\) and \(X_2\) are independent, say \(X_i \sim {\mathrm{Bern}}(0.5)\) independently. If we condition on some data \(Y\), will \(X_1\) and \(X_2\) always be independent under the posterior?